Modeling baseframe
This transversal team supports all the research tracks of the Chair by applying its modeling know-how to materials and (bio)processes. More precisely, this work aims to numerically support the scale-up. Thus, from small-scale experiments, designed in synergy between experimenters and modelers, it is possible to determine the key parameters of models that will be used to design large-scale systems. As the research areas cover a wide range of topics, the modeling baseframe is based on the mastery of many disciplines such as thermodynamics, continuum mechanics, coupled heat/mass transfers (even out of equilibrium), the modeling of biological systems and their coupling with physical models.
The modeling baseframe mobilizes skills in :
- Discrete modeling, to make the population behavior emerge from the individuals; continuous modeling, to apply our methods on large objects; homogenization, to make the link between the two previous approaches. Finally, we deploy machine learning methods to explore the complex multidimensional spaces represented by the biological datasets we generate.
- Scientific computing, based in particular on High Performance Computing (HPC) methods, to carry out large-scale simulations and virtual experiments. To this end, the Chair has a partnership with the URCA in order to have access to the resources of the ROMEO supercomputer.
Developed approaches
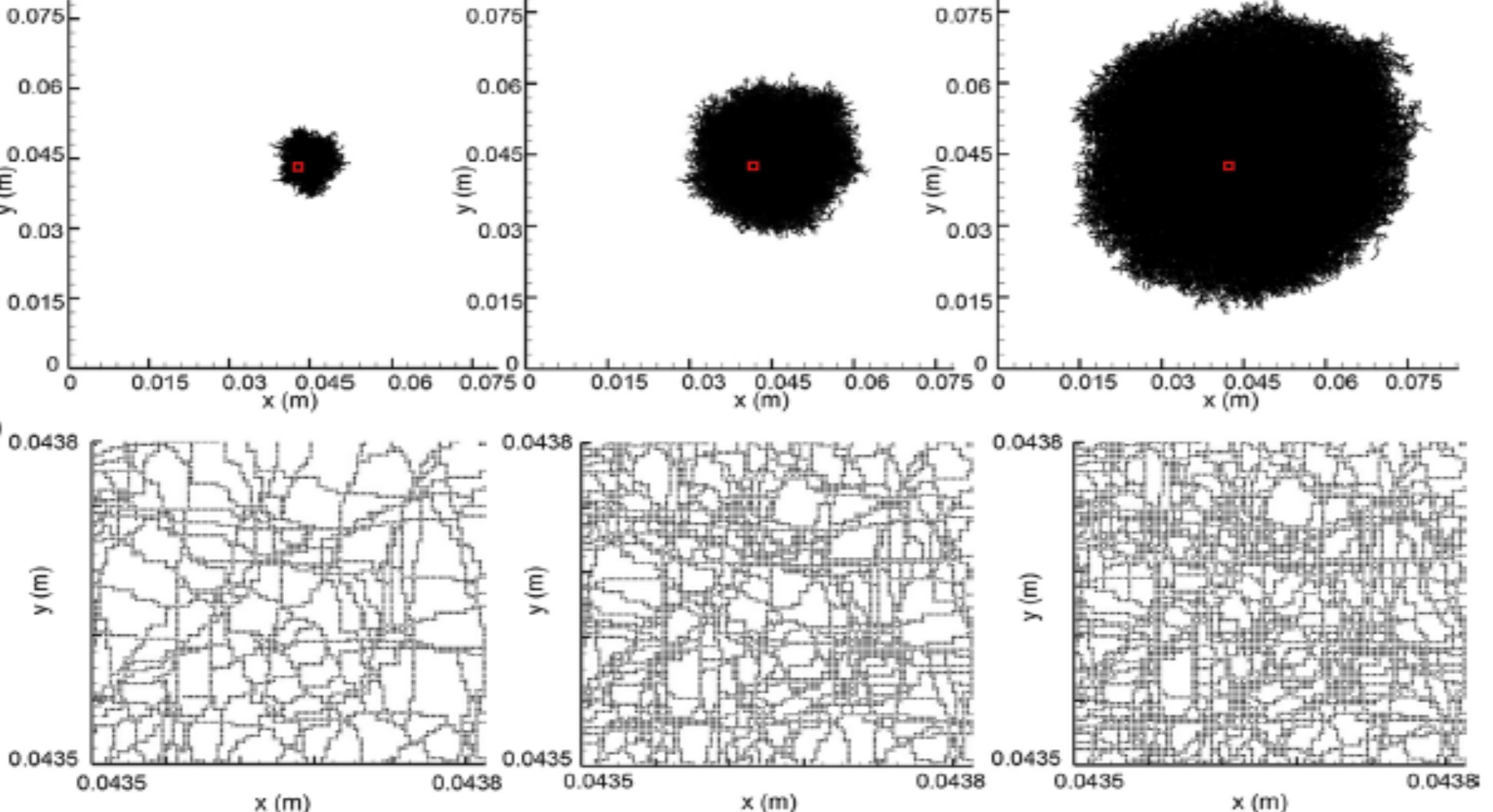
Discrete modeling
Discrete modeling is done at the level of individuals taken separately. Thus, each yeast, each microalgae has its own behavior. By considering a large number of individuals, potentially different, it is possible to obtain the behavior of the population. This approach is very rich in information, but usually limited to a local scale. However, thanks to HPC, it is possible to push the limits. For example, we are now able to fully reproduce a co-culture experiment on solid medium of yeast and microalgae.

Continuous modeling
The continuous medium approach modeling is concerned with macroscopic scale systems. It allows to consider processes/materials. For example, we use CFD (Computational Fluid Dynamics) to calculate fluid flows in bioreactors. Or Lattice-Boltzmann type approaches to simulate non-equilibrium heat transfer in materials. Our main field of interest is the evolution of moisture in lignocellulosic materials, on which the properties and longevity of the material depend. The use of HPC brings then the computational capacity to work on large space-time fields (4D) and thus to describe at best the objects of study.
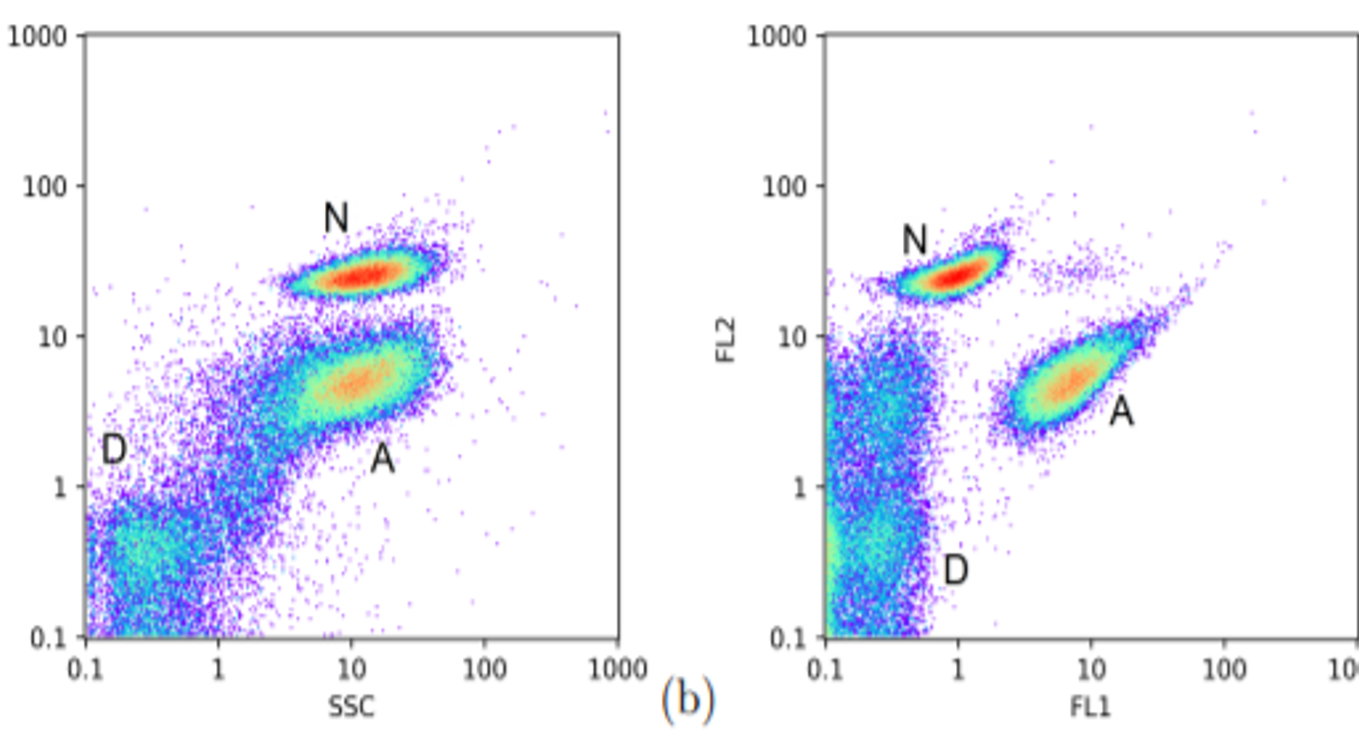
Machine learning
Today, biological datasets can be generated in large numbers. This is for example the case of genomic data. Although they are not as large, biological/biotechnological data have been greatly enriched over the last few years. For example, we use clustering methods to analyze our cytometric results (10,000 measurement points in 5 dimensions in a few minutes). Cell cultures are also complex environments with specific radiative properties. Thus, our spectral measurements (several thousand features) contain important information (quantity of cells, substrates, metabolite of interest) that we extract by machine learning.
Examples of achievements
• Computation of properties on real and virtual 3D morphologies (PredicTBiomat project) Continuous approach
• Design of a lighting device for photobioreactor, Discrete approach
• Coupled CFD & biological modeling of a photobioreactor, Continuous + discrete
• Modeling of microbial cultures on solid medium (microalgae, yeast, fungi) Discrete
• Modeling of mixed cultures: microalgae & yeasts, solid and liquid media Discrete, continuous
• Multiscale temperature/humidity calculations in building materials (SMART Reno project) continuous
• Upscaling of biogas and syngas purification solutions (Vitrhydrogen Project) Continuous
Resources
• Access to the ROMEO supercomputer
• Softwares: OpenFOAM, internally developed codes
• Programming languages: Fortran, R, Python, C++